Streaming (online) inference
This guide explains how to use the infer function for dynamic datasets. We show how RxInfer can continuously update beliefs asynchronously whenever a new observation arrives. We use a simple Beta-Bernoulli model as an example, which has been covered in the Getting Started section, however, these techniques can be applied to any model
Also read about Static Inference or checkout more complex examples.
Model specification
Also read the Model Specification section.
In online inference, we want to continuously update our prior beliefs about certain hidden states. To achieve this, we include extra arguments in our model specification to allow for dynamic prior changes:
using RxInfer
@model function beta_bernoulli_online(y, a, b)
θ ~ Beta(a, b)
y ~ Bernoulli(θ)
endIn this model, we assume we only have one observation y at a time, and the a and b parameters are not fixed to specific values but rather are arguments of the model itself.
Automatic prior update
Next, we want to enable RxInfer to automatically update the a and b parameters as soon as a new posterior for θ is available. To accomplish this, we utilize the @autoupdates macro.
beta_bernoulli_autoupdates = @autoupdates begin
# We want to update `a` and `b` to be equal to the parameters
# of the current posterior for `θ`
a, b = params(q(θ))
end@autoupdates begin
(a, b) = params(q(θ))
end
This specification instructs RxInfer to update a and b parameters automatically as as soon as a new posterior for θ is available. Read more about @autoupdates in the Autoupdates guide
Asynchronous data stream of observations
For demonstration purposes, we use a handcrafted stream of observations with the Rocket.jl library
using Rocket, Distributions, StableRNGs
hidden_θ = 1 / 3.1415
distribution = Bernoulli(hidden_θ)
rng = StableRNG(43)
datastream = RecentSubject(Bool)RecentSubject(Bool, Subject{Bool, AsapScheduler, AsapScheduler})The infer function expects the datastream to emit values in the form of the NamedTuples. To simplify this process, Rocket.jl exports labeled function. We also use the combineLatest function to convert a stream of Bools to a stream of Tuple{Bool}s. Read more about these function in the documentation to Rocket.jl.
observations = labeled(Val((:y, )), combineLatest(datastream))LabeledObservable(@NamedTuple{y::Bool}, Rocket.CombineLatestObservable{Tuple{Bool}, Tuple{Rocket.RecentSubjectInstance{Bool, Subject{Bool, AsapScheduler, AsapScheduler}}}, PushEach})Let's verify that our datastream does indeed produce NamedTuples
subscription = subscribe!(observations,
(new_observation) -> println("Got new observation ", new_observation, " 🎉")
)for i in 1:5
next!(datastream, rand(rng, distribution))
end
@test all(value -> haskey(value, :y) && (isone(value[:y]) || iszero(value[:y])), test_values) #hideGot new observation (y = false,) 🎉
Got new observation (y = true,) 🎉
Got new observation (y = false,) 🎉
Got new observation (y = true,) 🎉
Got new observation (y = false,) 🎉Nice! Our data stream produces events in a form of the NamedTuples, which is compatible with the infer function.
# It is important to keep track of the existing susbcriptions
# and unsubscribe to reduce the usage of computational resources
unsubscribe!(subscription)Instantiating the reactive inference engine
Now, we have everything ready to start running the inference with RxInfer on dynamic datasets with the infer function:
engine = infer(
model = beta_bernoulli_online(),
datastream = observations,
autoupdates = beta_bernoulli_autoupdates,
returnvars = (:θ, ),
initialization = @initialization(q(θ) = Beta(1, 1)),
autostart = false
)RxInferenceEngine:
Posteriors stream | enabled for (θ)
Free Energy stream | disabled
Posteriors history | unavailable
Free Energy history | unavailable
Enabled events | [ ]In the code above, there are several notable differences compared to running inference for static datasets. Firstly, we utilized the autoupdates argument as discussed previously. Secondly, we employed the @initialization macro to initialize the posterior over θ. This is necessary for the @autoupdates macro, as it needs to initialize the a and b parameters before the data becomes available. Thirdly, we set autostart = false to indicate that we do not want to immediately subscribe to the datastream, but rather do so manually later using the RxInfer.start function. The returnvars specification differs a little from Static Inference. In reactive inference, the returnvars = (:θ, ) must be a tuple of Symbols and specifies that we would be interested to get a stream of posteriors update for θ. The returnvars specification is optional and the inference engine will create reactive streams for all latent states if ommited.
RxInfer.RxInferenceEngine — TypeRxInferenceEngineThe return value of the infer function in case of streamlined inference.
Public fields
posteriors:DictorNamedTupleof 'random variable' - 'posterior stream' pairs. See thereturnvarsargument for theinfer.free_energy: (optional) A stream of Bethe Free Energy values per VMP iteration. See thefree_energyargument for theinfer.history: (optional) Saves history of previous marginal updates. See thehistoryvarsandkeephistoryarguments for theinfer.free_energy_history: (optional) Free energy history, averaged across variational iterations value for all observationsfree_energy_raw_history: (optional) Free energy history, returns returns computed values of all variational iterations for each data event (if available)free_energy_final_only_history: (optional) Free energy history, returns computed values of final variational iteration for each data event (if available)events: (optional) A stream of events send by the inference engine. See theeventsargument for theinfer.model:ProbabilisticModelobject reference.
Use the RxInfer.start(engine) function to subscribe on the datastream source and start the inference procedure. Use RxInfer.stop(engine) to unsubscribe from the datastream source and stop the inference procedure. Note, that it is not always possible to start/stop the inference procedure.
See also: infer, RxInferenceEvent, RxInfer.start, RxInfer.stop
RxInfer.start — Functionstart(engine::RxInferenceEngine)Starts the RxInferenceEngine by subscribing to the data source, instantiating free energy (if enabled) and starting the event loop. Use RxInfer.stop to stop the RxInferenceEngine. Note that it is not always possible to stop/restart the engine and this depends on the data source type.
See also: RxInfer.stop
RxInfer.stop — Functionstop(engine::RxInferenceEngine)Stops the RxInferenceEngine by unsubscribing to the data source, free energy (if enabled) and stopping the event loop. Use RxInfer.start to start the RxInferenceEngine again. Note that it is not always possible to stop/restart the engine and this depends on the data source type.
See also: RxInfer.start
Given the engine, we now can subscribe on the posterior updates:
θ_updates_subscription = subscribe!(engine.posteriors[:θ],
(new_posterior_for_θ) -> println("A new posterior for θ is ", new_posterior_for_θ, " 🤩")
)In this setting, we should get a message every time a new posterior is available for θ. Let's try to generate a new observation!
next!(datastream, rand(rng, distribution))Hmm, nothing happened...? Oh, we forgot to start the engine with the RxInfer.start function. Let's do that now:
RxInfer.start(engine)A new posterior for θ is Beta{Float64}(α=1.0, β=2.0) 🤩Ah, as soon as we start our engine, we receive the posterior for θ. This occurred because we initialized our stream as RecentSubject, which retains the most recent value and emits it upon subscription. Our engine automatically subscribed to the observations and obtained the most recent value, initiating inference. Let's see if we can add more observations:
next!(datastream, rand(rng, distribution))A new posterior for θ is Beta{Float64}(α=1.0, β=3.0) 🤩Great! We got another posterior! Let's try a few more observations:
for i in 1:5
next!(datastream, rand(rng, distribution))
endA new posterior for θ is Beta{Float64}(α=2.0, β=3.0) 🤩
A new posterior for θ is Beta{Float64}(α=2.0, β=4.0) 🤩
A new posterior for θ is Beta{Float64}(α=2.0, β=5.0) 🤩
A new posterior for θ is Beta{Float64}(α=2.0, β=6.0) 🤩
A new posterior for θ is Beta{Float64}(α=2.0, β=7.0) 🤩As demonstrated, the reactive engine reacts to new observations and performs inference as soon as a new observation is available. But what if we want to maintain a history of posteriors? The infer function supports the historyvars and keephistory arguments precisely for that purpose. In the next section we reinstantiate our engine, with the keephistory argument enabled, but first, we must shutdown the previous engine and unsubscribe from its posteriors:
RxInfer.stop(engine)
unsubscribe!(θ_updates_subscription)Keeping the history of posteriors
To retain the history of posteriors within the engine, we can utilize the keephistory and historyvars arguments. The keephistory parameter specifies the length of the circular buffer for storing the history of posterior updates, while historyvars determines what variables to save in the history and how often to save them (e.g., every iteration or only at the end of iterations).
engine = infer(
model = beta_bernoulli_online(),
datastream = observations,
autoupdates = beta_bernoulli_autoupdates,
initialization = @initialization(q(θ) = Beta(1, 1)),
keephistory = 100,
historyvars = (θ = KeepLast(), ),
autostart = true
)RxInferenceEngine:
Posteriors stream | enabled for (θ)
Free Energy stream | disabled
Posteriors history | available for (θ)
Free Energy history | unavailable
Enabled events | [ ]In the example above, we specified that we want to store at most 100 posteriors for θ, and KeepLast() indicates that we are only interested in the final value of θ and not in intermediate values during variational iterations. We also specified the autostart = true to start the engine automatically without need for RxInfer.start and RxInfer.stop.
In this model, we do not utilize the iterations argument, indicating that we perform a single VMP iteration. If multiple iterations were employed, engine.posteriors[:θ] would emit every intermediate value.
Now, we can feed some more observations to the datastream:
for i in 1:5
next!(datastream, rand(rng, distribution))
endAnd inspect the engine.history[:θ] buffer:
engine.history[:θ]6-element DataStructures.CircularBuffer{Any}:
Beta{Float64}(α=1.0, β=2.0)
Beta{Float64}(α=2.0, β=2.0)
Beta{Float64}(α=3.0, β=2.0)
Beta{Float64}(α=3.0, β=3.0)
Beta{Float64}(α=3.0, β=4.0)
Beta{Float64}(α=3.0, β=5.0)As we can see the buffer correctly saved the posteriors in the .history buffer.
We have 6 entries, despite having only 5 new observations. As mentioned earlier, this occurs because we initialized our datastream as a RecentSubject, which retains the most recent observation and emits it each time a new subscription occurs.
Visualizing the history of posterior estimation
Let's feed more observation and visualize how the posterior changes over time:
for i in 1:94
next!(datastream, rand(rng, distribution))
endTo visualize the history of posteriors we use the @gif macro from the Plots package:
using Plots
@gif for posterior in engine.history[:θ]
rθ = range(0, 1, length = 1000)
pθ = plot(rθ, (x) -> pdf(posterior, x), fillalpha=0.3, fillrange = 0, label="P(θ|y)", c=3)
pθ = vline!(pθ, [ hidden_θ ], label = "Real value of θ")
plot(pθ)
end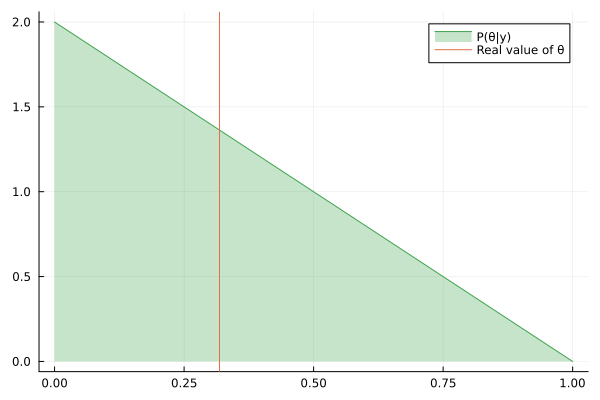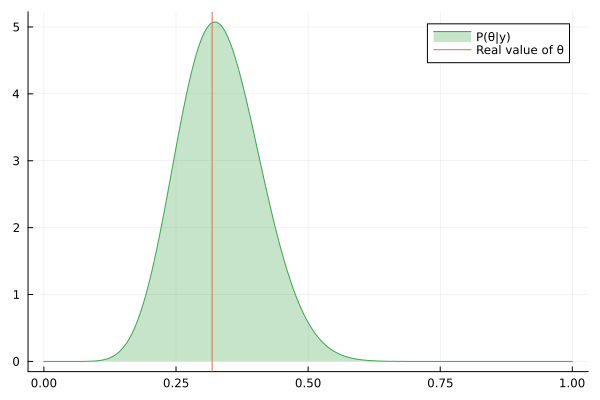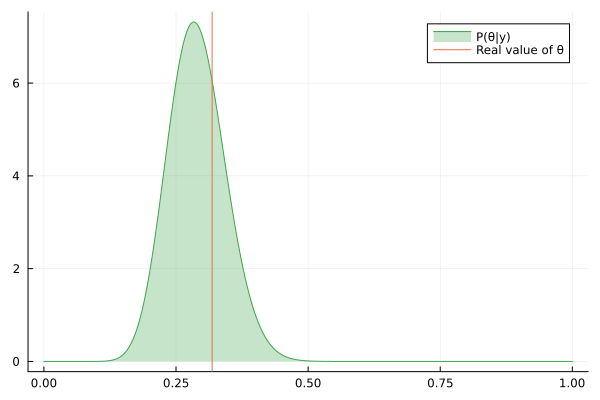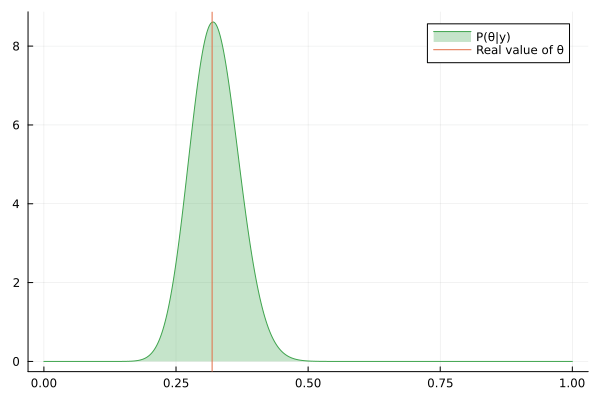
We can keep feeding data to our datastream, but only last 100 posteriors will be saved in the history buffer:
for i in 1:200
next!(datastream, rand(rng, distribution))
end
@gif for posterior in engine.history[:θ]
rθ = range(0, 1, length = 1000)
pθ = plot(rθ, (x) -> pdf(posterior, x), fillalpha=0.3, fillrange = 0, label="P(θ|y)", c=3)
pθ = vline!(pθ, [ hidden_θ ], label = "Real value of θ")
plot(pθ)
end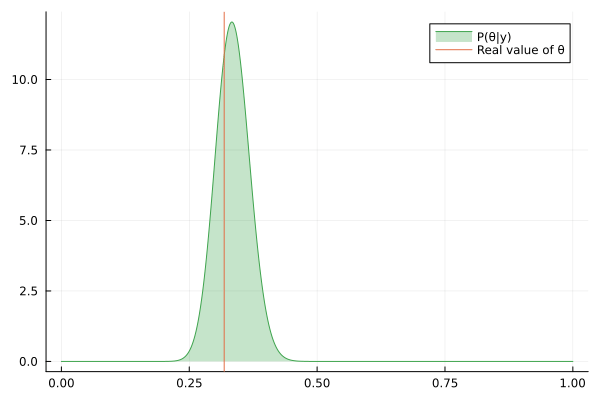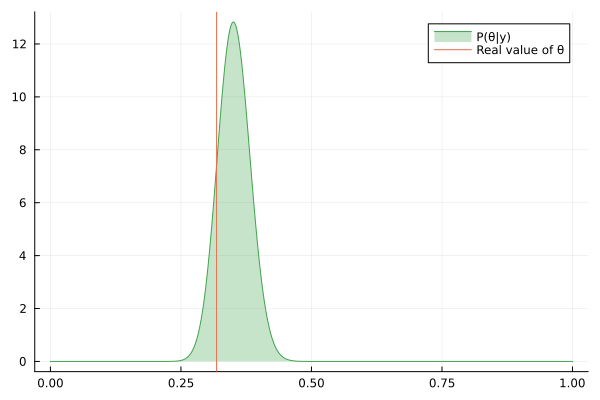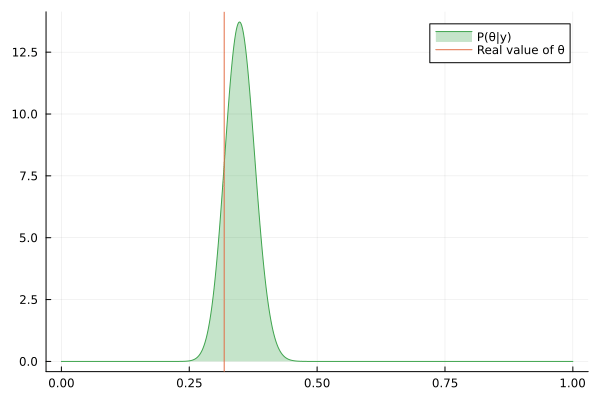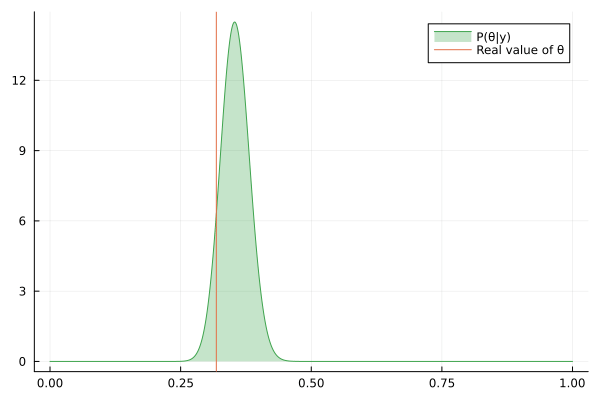
It is also possible to visualize the inference estimation continously with manual subscription to engine.posteriors[:θ].
As previously it is important to shutdown the inference engine when it becomes unnecessary:
RxInfer.stop(engine)Subscribing on the stream of free energy
To obtain a continuous stream of updates for the Bethe Free Energy, we need to initialize the engine with the free_energy argument set to true:
engine = infer(
model = beta_bernoulli_online(),
datastream = observations,
autoupdates = beta_bernoulli_autoupdates,
initialization = @initialization(q(θ) = Beta(1, 1)),
keephistory = 5,
autostart = true,
free_energy = true
)RxInferenceEngine:
Posteriors stream | enabled for (θ)
Free Energy stream | enabled
Posteriors history | available for (θ)
Free Energy history | available
Enabled events | [ ]It's important to use the keephistory argument alongside the free_energy argument because setting free_energy = true also maintains an internal circular buffer to track its previous updates.
free_energy_subscription = subscribe!(engine.free_energy,
(bfe_value) -> println("New value of Bethe Free Energy has been computed ", bfe_value, " 👩🔬")
)New value of Bethe Free Energy has been computed 0.6931471805599452 👩🔬Let's emit more observations:
for i in 1:5
next!(datastream, rand(rng, distribution))
endNew value of Bethe Free Energy has been computed 0.4054651081081643 👩🔬
New value of Bethe Free Energy has been computed 0.28768207245178123 👩🔬
New value of Bethe Free Energy has been computed 1.6094379124340998 👩🔬
New value of Bethe Free Energy has been computed 1.0986122886681102 👩🔬
New value of Bethe Free Energy has been computed 0.5596157879354218 👩🔬In this particular example, we do not perform any variational iterations and do not use any variational constraints, hence, the inference is exact. In this case the BFE values are equal to the minus log-evidence of the model given new observation. We can also track history of Bethe Free Energy values with the following fields of the engine:
free_energy_history: free energy history, averaged across variational iterations value for all observationsfree_energy_raw_history: free energy history, returns returns computed values of all variational iterations for each data event (if available)free_energy_final_only_history: free energy history, returns computed values of final variational iteration for each data event (if available)
engine.free_energy_history1-element Vector{Real}:
0.7921626339195154engine.free_energy_raw_history5-element Vector{Real}:
0.4054651081081643
0.28768207245178123
1.6094379124340998
1.0986122886681102
0.5596157879354218engine.free_energy_final_only_history5-element Vector{Real}:
0.4054651081081643
0.28768207245178123
1.6094379124340998
1.0986122886681102
0.5596157879354218# Stop the engine when not needed as usual
RxInfer.stop(engine)
unsubscribe!(free_energy_subscription)As has been mentioned, in this particular example we do not perform variational iterations, hence, there is little different between different representations of the BFE history buffers. However, when performing variational inference with the iterations argument, those buffers will be different. To demonstrate this difference let's build a slightly more complex model with variational constraints:
@model function iid_normal(y, mean_μ, var_μ, shape_τ, rate_τ)
μ ~ Normal(mean = mean_μ, var = var_μ)
τ ~ Gamma(shape = shape_τ, rate = rate_τ)
y ~ Normal(mean = μ, precision = τ)
end
iid_normal_constraints = @constraints begin
q(μ, τ) = q(μ)q(τ)
end
iid_normal_autoupdates = @autoupdates begin
mean_μ = mean(q(μ))
var_μ = var(q(μ))
shape_τ = shape(q(τ))
rate_τ = rate(q(τ))
end
iid_normal_hidden_μ = 3.1415
iid_normal_hidden_τ = 0.0271
iid_normal_distribution = NormalMeanPrecision(iid_normal_hidden_μ, iid_normal_hidden_τ)
iid_normal_rng = StableRNG(123)
iid_normal_datastream = RecentSubject(Float64)
iid_normal_observations = labeled(Val((:y, )), combineLatest(iid_normal_datastream))
iid_normal_initialization = @initialization begin
q(μ) = NormalMeanPrecision(0.0, 0.001)
q(τ) = GammaShapeRate(10.0, 10.0)
end
iid_normal_engine = infer(
model = iid_normal(),
datastream = iid_normal_observations,
autoupdates = iid_normal_autoupdates,
constraints = iid_normal_constraints,
initialization = iid_normal_initialization,
historyvars = (
μ = KeepLast(),
τ = KeepLast(),
),
keephistory = 100,
iterations = 10,
free_energy = true,
autostart = true
)RxInferenceEngine:
Posteriors stream | enabled for (μ, τ)
Free Energy stream | enabled
Posteriors history | available for (μ, τ)
Free Energy history | available
Enabled events | [ ]The notable differences with the previous example is the use of the constraints and iterations arguments. Read more about constraints in the Constraints Specification section of the documentation. We have also indicated in the historyvars that we want to keep track of posteriors only from the last variational iteration in the history buffer.
Now we can feed some observations to the datastream:
for i in 1:100
next!(iid_normal_datastream, rand(iid_normal_rng, iid_normal_distribution))
endLet's inspect the differences in the free_energy buffers:
iid_normal_engine.free_energy_history10-element Vector{Real}:
3.6043166919134983
3.583392877186756
3.5826032453113443
3.5825781243744648
3.5825773364749796
3.5825773108212466
3.582577309930648
3.5825773098970286
3.582577309895637
3.5825773098955747iid_normal_engine.free_energy_raw_history1000-element Vector{Real}:
4.400915492231585
4.400915491646703
4.400915491645383
4.400915491645376
4.400915491645378
4.40091549164538
4.400915491645378
4.400915491645379
4.400915491645382
4.400915491645381
⋮
3.3459102809749774
3.345910280974934
3.3459102809749397
3.3459102809749406
3.3459102809749406
3.3459102809749406
3.3459102809749406
3.3459102809749406
3.3459102809749406iid_normal_engine.free_energy_final_only_history100-element Vector{Real}:
4.400915491645381
6.750221908665356
15.238854420361326
1.732109425738347
1.708353615319435
5.532943388368783
3.7219642469057916
3.5831347993392586
4.483661427185016
6.051352052233973
⋮
3.0121619504111528
3.2849460863247915
2.794188080870387
4.190015761850561
3.2727955937808066
5.350758196381457
2.7547733362534608
2.7673806065614217
3.3459102809749406We can also visualize different representations:
plot(iid_normal_engine.free_energy_history, label = "Bethe Free Energy (averaged)")In general, the averaged Bethe Free Energy values must decrease and converge to a stable point.
plot(iid_normal_engine.free_energy_raw_history, label = "Bethe Free Energy (raw)")plot(iid_normal_engine.free_energy_final_only_history, label = "Bethe Free Energy (last per observation)")As we can see, in the case of the variational iterations those buffers are quite different and represent different representations of the same Bethe Free Energy stream (which corresponds to the .free_energy_raw_history). As a sanity check, we could also visualize the history of our posterior estimations in the same way as we did for a simpler previous example:
@gif for (μ_posterior, τ_posterior) in zip(iid_normal_engine.history[:μ], iid_normal_engine.history[:τ])
rμ = range(0, 10, length = 1000)
rτ = range(0, 1, length = 1000)
pμ = plot(rμ, (x) -> pdf(μ_posterior, x), fillalpha=0.3, fillrange = 0, label="P(μ|y)", c=3)
pμ = vline!(pμ, [ iid_normal_hidden_μ ], label = "Real value of μ")
pτ = plot(rτ, (x) -> pdf(τ_posterior, x), fillalpha=0.3, fillrange = 0, label="P(τ|y)", c=3)
pτ = vline!(pτ, [ iid_normal_hidden_τ ], label = "Real value of τ")
plot(pμ, pτ, layout = @layout([ a; b ]))
end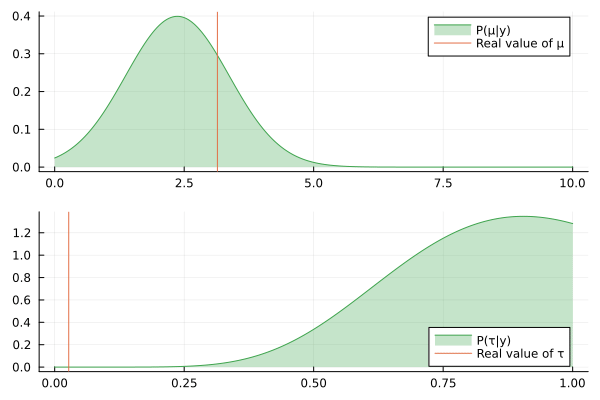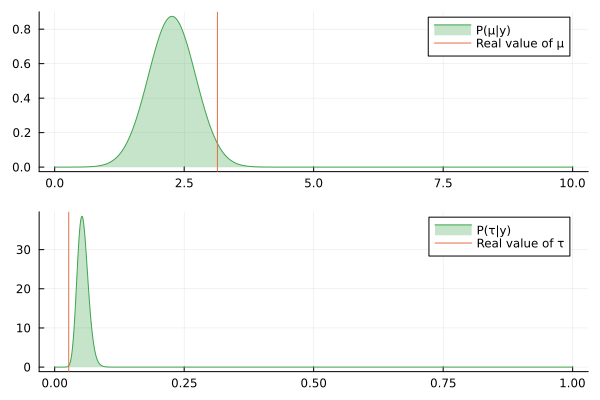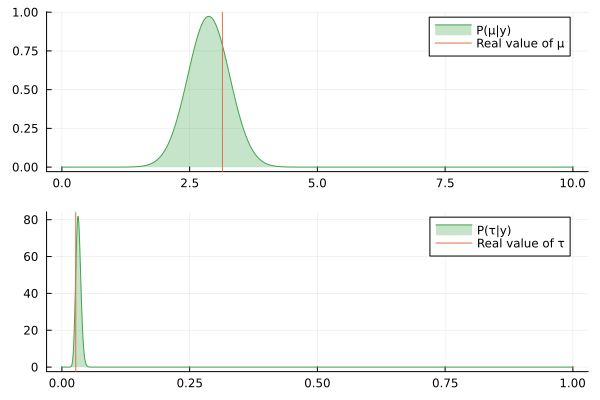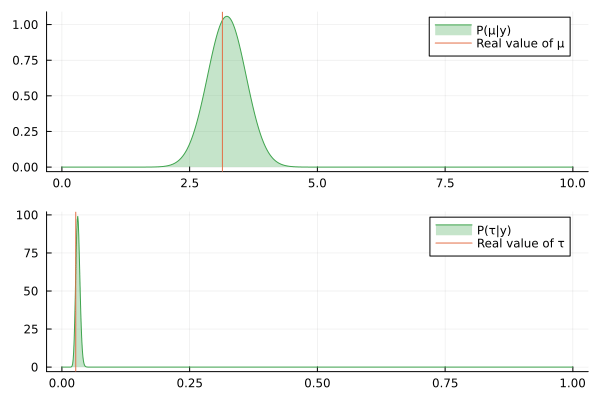
Nice, the history of the estimated posteriors aligns well with the real (hidden) values of the underlying parameters.
Callbacks
The RxInferenceEngine has its own lifecycle. The callbacks differ a little bit from Using callbacks with Static Inference. Here are available callbacks that can be used together with the streaming inference:
before_model_creation()Calls before the model is going to be created, does not accept any arguments.
after_model_creation(model::ProbabilisticModel)Calls right after the model has been created, accepts a single argument, the model.
before_autostart(engine::RxInferenceEngine)Calls before the RxInfer.start() function, if autostart is set to true.
after_autostart(engine::RxInferenceEngine)Calls after the RxInfer.start() function, if autostart is set to true.
Here is an example usage of the outlined callbacks:
function before_model_creation()
println("The model is about to be created")
end
function after_model_creation(model::ProbabilisticModel)
println("The model has been created")
println(" The number of factor nodes is: ", length(RxInfer.getfactornodes(model)))
println(" The number of latent states is: ", length(RxInfer.getrandomvars(model)))
println(" The number of data points is: ", length(RxInfer.getdatavars(model)))
println(" The number of constants is: ", length(RxInfer.getconstantvars(model)))
end
function before_autostart(engine::RxInferenceEngine)
println("The reactive inference engine is about to start")
end
function after_autostart(engine::RxInferenceEngine)
println("The reactive inference engine has been started")
end
engine = infer(
model = beta_bernoulli_online(),
datastream = observations,
autoupdates = beta_bernoulli_autoupdates,
initialization = @initialization(q(θ) = Beta(1, 1)),
keephistory = 5,
autostart = true,
free_energy = true,
callbacks = (
before_model_creation = before_model_creation,
after_model_creation = after_model_creation,
before_autostart = before_autostart,
after_autostart = after_autostart
)
)The model is about to be created
The model has been created
The number of factor nodes is: 2
The number of latent states is: 1
The number of data points is: 3
The number of constants is: 0
The reactive inference engine is about to start
The reactive inference engine has been startedEvent loop
In constrast to Static Inference, the streaming version of the infer function does not provide callbacks such as on_marginal_update, since it is possible to subscribe directly on those updates with the engine.posteriors field. However, the reactive inference engine provides an ability to listen to its internal event loop, that also includes "pre" and "post" events for posterior updates.
RxInfer.RxInferenceEvent — TypeRxInferenceEvent{T, D}The RxInferenceEngine sends events in a form of the RxInferenceEvent structure. T represents the type of an event, D represents the type of a data associated with the event. The type of data depends on the type of an event, but usually represents a tuple, which can be unrolled automatically with the Julia's splitting syntax, e.g. model, iteration = event. See the documentation of the rxinference function for possible event types and their associated data types.
The events system itself uses the Rocket.jl library API. For example, one may create a custom event listener in the following way:
using Rocket
struct MyEventListener <: Rocket.Actor{RxInferenceEvent}
# ... extra fields
end
function Rocket.on_next!(listener::MyEventListener, event::RxInferenceEvent{ :after_iteration })
model, iteration = event
println("Iteration $(iteration) has been finished.")
end
function Rocket.on_error!(listener::MyEventListener, err)
# ...
end
function Rocket.on_complete!(listener::MyEventListener)
# ...
end
and later on:
engine = infer(events = Val((:after_iteration, )), ...)
subscription = subscribe!(engine.events, MyEventListener(...))See also: infer, RxInferenceEngine
Let's build a simple example by implementing our own event listener that does not do anything complex but simply prints some debugging information.
struct MyEventListener <: Rocket.Actor{RxInferenceEvent}
# ... extra fields
endThe available events are
:before_startEmits right before starting the engine with the RxInfer.start function. The data is (engine::RxInferenceEngine, )
function Rocket.on_next!(listener::MyEventListener, event::RxInferenceEvent{ :before_start })
(engine, ) = event
println("The engine is about to start.")
end:after_startEmits right after starting the engine with the RxInfer.start function. The data is (engine::RxInferenceEngine, )
function Rocket.on_next!(listener::MyEventListener, event::RxInferenceEvent{ :after_start })
(engine, ) = event
println("The engine has been started.")
end:before_stopEmits right before stopping the engine with the RxInfer.stop function. The data is (engine::RxInferenceEngine, )
function Rocket.on_next!(listener::MyEventListener, event::RxInferenceEvent{ :before_stop })
(engine, ) = event
println("The engine is about to be stopped.")
end:after_stopEmits right after stopping the engine with the RxInfer.stop function. The data is (engine::RxInferenceEngine, )
function Rocket.on_next!(listener::MyEventListener, event::RxInferenceEvent{ :after_stop })
(engine, ) = event
println("The engine has been stopped.")
end:on_new_dataEmits right before processing new data point. The data is (model::ProbabilisticModel, data)
function Rocket.on_next!(listener::MyEventListener, event::RxInferenceEvent{ :on_new_data })
(model, data) = event
println("The new data point has been received: ", data)
end:before_iterationEmits right before starting new variational iteration. The data is (model::ProbabilisticModel, iteration::Int)
function Rocket.on_next!(listener::MyEventListener, event::RxInferenceEvent{ :before_iteration })
(model, iteration) = event
println("Starting new variational iteration #", iteration)
end:before_auto_updateEmits right before executing the @autoupdates. The data is (model::ProbabilisticModel, iteration::Int, autoupdates)
function Rocket.on_next!(listener::MyEventListener, event::RxInferenceEvent{ :before_auto_update })
(model, iteration, autoupdates) = event
println("Before processing autoupdates")
end:after_auto_updateEmits right after executing the @autoupdates. The data is (model::ProbabilisticModel, iteration::Int, autoupdates)
function Rocket.on_next!(listener::MyEventListener, event::RxInferenceEvent{ :after_auto_update })
(model, iteration, autoupdates) = event
println("After processing autoupdates")
end:before_data_updateEmits right before feeding the model with the new data. The data is (model::ProbabilisticModel, iteration::Int, data)
function Rocket.on_next!(listener::MyEventListener, event::RxInferenceEvent{ :before_data_update })
(model, iteration, data) = event
println("Before processing new data ", data)
end:after_data_updateEmits right after feeding the model with the new data. The data is (model::ProbabilisticModel, iteration::Int, data)
function Rocket.on_next!(listener::MyEventListener, event::RxInferenceEvent{ :after_data_update })
(model, iteration, data) = event
println("After processing new data ", data)
end:after_iterationEmits right after finishing a variational iteration. The data is (model::ProbabilisticModel, iteration::Int)
function Rocket.on_next!(listener::MyEventListener, event::RxInferenceEvent{ :after_iteration })
(model, iteration) = event
println("Finishing the variational iteration #", iteration)
end:before_history_saveEmits right before saving the history (if requested). The data is (model::ProbabilisticModel, )
function Rocket.on_next!(listener::MyEventListener, event::RxInferenceEvent{ :before_history_save })
(model, ) = event
println("Before saving the history")
end:after_history_saveEmits right after saving the history (if requested). The data is (model::ProbabilisticModel, )
function Rocket.on_next!(listener::MyEventListener, event::RxInferenceEvent{ :after_history_save })
(model, ) = event
println("After saving the history")
end:on_tickEmits right after finishing processing the new observations and completing the inference step. The data is (model::ProbabilisticModel, )
function Rocket.on_next!(listener::MyEventListener, event::RxInferenceEvent{ :on_tick })
(model, ) = event
println("Finishing the inference for the new observations")
end:on_errorEmits if an error occurs in the inference engine. The data is (model::ProbabilisticModel, err::Any)
function Rocket.on_next!(listener::MyEventListener, event::RxInferenceEvent{ :on_error })
(model, err) = event
println("An error occured during the inference procedure: ", err)
end:on_completeEmits when the datastream completes. The data is (model::ProbabilisticModel, )
function Rocket.on_next!(listener::MyEventListener, event::RxInferenceEvent{ :on_complete })
(model, ) = event
println("The data stream completed. The inference has been finished.")
endLet's use our event listener with the infer function:
engine = infer(
model = beta_bernoulli_online(),
datastream = observations,
autoupdates = beta_bernoulli_autoupdates,
initialization = @initialization(q(θ) = Beta(1, 1)),
keephistory = 5,
iterations = 2,
autostart = false,
free_energy = true,
events = Val((
:before_start,
:after_start,
:before_stop,
:after_stop,
:on_new_data,
:before_iteration,
:before_auto_update,
:after_auto_update,
:before_data_update,
:after_data_update,
:after_iteration,
:before_history_save,
:after_history_save,
:on_tick,
:on_error,
:on_complete
))
)RxInferenceEngine:
Posteriors stream | enabled for (θ)
Free Energy stream | enabled
Posteriors history | available for (θ)
Free Energy history | available
Enabled events | [ before_start, after_start, before_stop, after_stop, on_new_data, before_iteration, before_auto_update, after_auto_update, before_data_update, after_data_update, after_iteration, before_history_save, after_history_save, on_tick, on_error, on_complete ]After we have created the engine, we can subscribe on events and RxInfer.start the engine:
events_subscription = subscribe!(engine.events, MyEventListener())
RxInfer.start(engine)The engine is about to start.
The new data point has been received: (y = false,)
Starting new variational iteration #1
Before processing autoupdates
After processing autoupdates
Before processing new data (y = false,)
After processing new data (y = false,)
Finishing the variational iteration #1
Starting new variational iteration #2
Before processing autoupdates
After processing autoupdates
Before processing new data (y = false,)
After processing new data (y = false,)
Finishing the variational iteration #2
Before saving the history
After saving the history
Finishing the inference for the new observations
The engine has been started.The event loop stays idle without new observation and runs again when a new observation becomes available:
next!(datastream, rand(rng, distribution))The new data point has been received: (y = false,)
Starting new variational iteration #1
Before processing autoupdates
After processing autoupdates
Before processing new data (y = false,)
After processing new data (y = false,)
Finishing the variational iteration #1
Starting new variational iteration #2
Before processing autoupdates
After processing autoupdates
Before processing new data (y = false,)
After processing new data (y = false,)
Finishing the variational iteration #2
Before saving the history
After saving the history
Finishing the inference for the new observationsLet's complete the datastream
complete!(datastream)The data stream completed. The inference has been finished.In this case, it is not necessary to RxInfer.stop the engine, because it will be stopped automatically.
RxInfer.stop(engine)┌ Warning: The engine has been completed or errored. Cannot stop an exhausted engine.
└ @ RxInfer ~/work/RxInfer.jl/RxInfer.jl/src/inference/streaming.jl:240The :before_stop and :after_stop events are not emmited in case of the datastream completion. Use the :on_complete instead.
Using data keyword argument with streaming inference
The streaming version does support static datasets as well. Internally, it converts it to a datastream, that emits all observations in a sequntial order without any delay. As an example:
staticdata = rand(rng, distribution, 1_000)1000-element Vector{Bool}:
0
1
0
0
1
0
0
0
0
0
⋮
0
0
0
0
1
1
0
0
0Use the data keyword argument instead of the datastream to pass the static data.
engine = infer(
model = beta_bernoulli_online(),
data = (y = staticdata, ),
autoupdates = beta_bernoulli_autoupdates,
initialization = @initialization(q(θ) = Beta(1, 1)),
keephistory = 1000,
autostart = true,
free_energy = true,
)RxInferenceEngine:
Posteriors stream | enabled for (θ)
Free Energy stream | enabled
Posteriors history | available for (θ)
Free Energy history | available
Enabled events | [ ]engine.history[:θ]1000-element DataStructures.CircularBuffer{Any}:
Beta{Float64}(α=1.0, β=2.0)
Beta{Float64}(α=2.0, β=2.0)
Beta{Float64}(α=2.0, β=3.0)
Beta{Float64}(α=2.0, β=4.0)
Beta{Float64}(α=3.0, β=4.0)
Beta{Float64}(α=3.0, β=5.0)
Beta{Float64}(α=3.0, β=6.0)
Beta{Float64}(α=3.0, β=7.0)
Beta{Float64}(α=3.0, β=8.0)
Beta{Float64}(α=3.0, β=9.0)
⋮
Beta{Float64}(α=322.0, β=672.0)
Beta{Float64}(α=322.0, β=673.0)
Beta{Float64}(α=322.0, β=674.0)
Beta{Float64}(α=322.0, β=675.0)
Beta{Float64}(α=323.0, β=675.0)
Beta{Float64}(α=324.0, β=675.0)
Beta{Float64}(α=324.0, β=676.0)
Beta{Float64}(α=324.0, β=677.0)
Beta{Float64}(α=324.0, β=678.0)@gif for posterior in engine.history[:θ]
rθ = range(0, 1, length = 1000)
pθ = plot(rθ, (x) -> pdf(posterior, x), fillalpha=0.3, fillrange = 0, label="P(θ|y)", c=3)
pθ = vline!(pθ, [ hidden_θ ], label = "Real value of θ")
plot(pθ)
end
Where to go next?
This guide covered some fundamental usages of the infer function in the context of streamline inference, but did not cover all the available keyword arguments of the function. Read more explanation about the other keyword arguments in the Overview section or check out the Static Inference section. Also check out more complex examples.